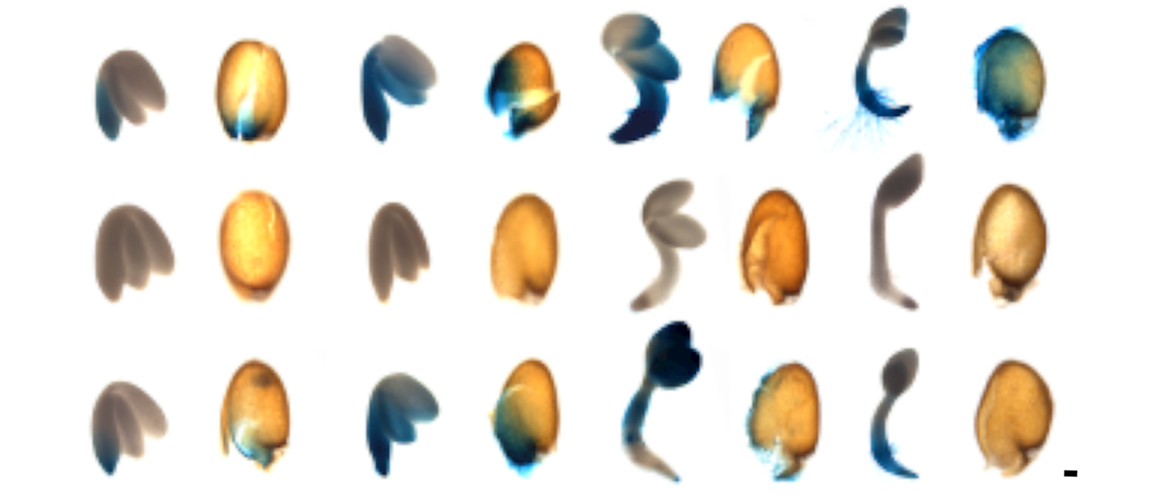Current research focus
Our work on understanding the C4 pathway makes use of model organisms as well as natural variation, and classical molecular biology approaches as well as emerging technologies in bioinformatics and computational processing.

Comparative analysis of C3 and C4 gene expression
With Andreas Weber’s lab we initially conducted analysis of RNA populations in whole leaves of C3 and C4 Cleome species. We are extending this to undertake large-scale analysis of many more lineages, of leaf maturation, and of mesophyll and bundle sheath transcriptomes. To fully understand these large dataset, significant investment in development of computational approaches is required!
Images: top and left: traced leaf venation patterns of C3 vs. C4 plants.
The ancestral role of proteins used in C4 photosynthesis
Despite having well-characterised functions in C4 species, many of the core enzymes of the C4 pathway have poorly understood roles in C3 plants. We are analyzing the roles that they play through analysis of when and where they are expressed, as well as the use of insertional mutants and overexpression, followed by in depth photosynthetic and metabolic phenotyping. Many of these proteins are members of multi-gene families, and belong to redundant networks, and so the analysis is often intellectually challenging.
Images: top and left: developing Arabidopsis seedlings expressing a marker

The C4 rice project
Our focus has been generating resources that allow us to test the extent to which the basic C4 cycle can be installed in rice. This has involved analysis of the extent to which C4 genes from maize are expressed in a cell specific manner in rice, and also investigating potential regulators of these genes. Clearly, this is a long-term, incredibly challenging project, but we are finding that we learn a lot about the fundamentals of C4 photosynthesis along the way.
Images: top: C3 rice (foreground) growing alongside C4 Echinochloa glabrescens (mid) and maize (rear). The increased efficiency of C4 enables the C4 plants to grow faster with the same resources. left: rice growing in paddy conditions.
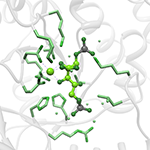
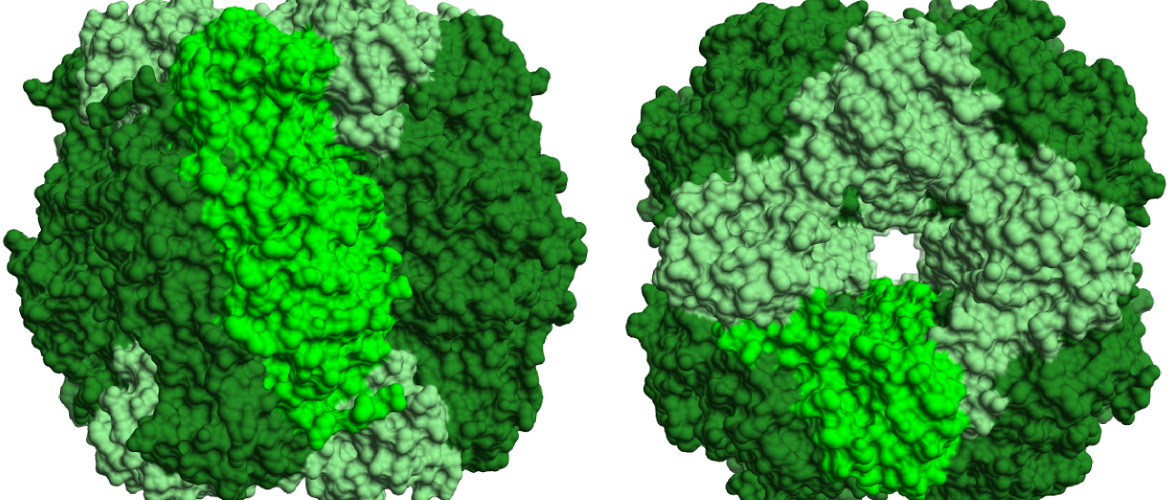
Using synthetic approaches to increase CO2 supply to RuBisCO
For many decades, work has been conducted that aims to increase the efficiency of RuBisCO by modifying its catalytic characteristics. We are taking an alternate approach inspired by Synthetic Biology in order to investigate whether artificial protein scaffolds can be used to improve the efficiency of photosynthesis. We hope that we can use this as proof of concept for engineering plant metabolism in general.
Images: top: structure of RuBisCO complex consisting of 8 large and and 8 small subunits. left: RuBP binding the active site of the RuBisCO large subunit.

Overcoming the domestication bottleneck in rice
A significant amount of work has been undertaken in which individual wild relatives of rice have been used as sources for useful traits in domesticated rice. In this project we are broadening this concept to source variation present in all the AA rice genomes, and increase it further using the multi advance generation intercrossing approach. Back crossing into O. sativa allows useful traits to transferred into elite germplasm. The wide crossing, and the subsequent analysis of gene expression and genomic structures represent significant technical and computational challenges.
Images: top and left: wild relatives of rice showing the broad morphological spectrum in the genus. This project aims to harness that diversity to improve cultivated rice.
Past projects and funding
- Cleome as a model for C4 photosynthesis (The Leverhulme Trust)
- The role and regulation of PPDK in C3 species (BBSRC Responsive mode)
- The role of C4 acid decarboxylases in Arabidopsis (BBSRC Responsive mode)
- Photosynthesis in discrete cell types. (BBSRC Sir David Phillips Research Fellowship)
Collaborations:
- With Dr Alex Webb. The circadian clock and photosynthesis. (BBSRC Responsive mode)
- With Dr Mark Tester and Emmanuel Guiderdoni. Rice enhancer traps (BBSRC Responsive mode)